Vertebrates are modified segmented worms; that is, their body plan is made up of sequentially repeated units, most apparent in skeletal structures like the vertebrae.
Arthropods are also modified segmented worms. Look at a larval fly, for instance, and you can see they are made up of rings stacked together.
So here's a simple and obvious question: can we infer that the last common ancestor of vertebrates and arthropods was also a segmented worm? That is, is segmentation a common ancestral trait, or did arthropods and vertebrates invent it independently? At first thought, you might assume they are: it's a complex trait shared by two taxa, so the simplest assumption is that both groups inherited it from their common ancestor (making it a synapomorphy), but there are also substantial differences in the mechanism of segmentation, so it's possible that this trait wasn't present in the common ancestor (making it a homoplasy).
Which is it? And the answer is…we don't know! There's a great deal of sympathy for synapomorphy, driven largely by a molecular bias — we see that a lot of the genes involved in the process in both vertebrates and arthropods are shared. There is a whole family of Notch-related cyclic genes, for instance, that turn out to be important in both, but the catch is that Notch is a gene that gets recruited in all kinds of processes — it's part of a handy developmental module for defining borders. So its presence doesn't automatically imply homology.
And then there's the whole problem of segmentation looking so different in flies (weird, highly derived arthropods) and vertebrates.
Flies build their segments almost all at once. In the fly embryo, there is first a broad gradient of position information, then a set of genes called the gap genes are switched on to define broad zones in the animal, and then, finally, the segmentation genes read the pattern of the gap genes and interact with each other to partition the fly into segments. It means segmentation is fast: all 14 segments form in one short interval, nearly simultaneously.
There's another complication. The segmentation genes in flies are numerous and elaborate, and in particular, they set up a peculiar pattern of alternating periodicity using genes called pair-rule genes. That means that there is one set of genes active in all of the odd-numbered segments (eve, or even-skipped, for instance), and a different set active in all of the even-numbered segments (ftz, or fushitarazu). In addition, there's a regular platoon of other pair-rule genes like odd and prd and hairy that are expressed in alternating segmental domains.
What it means is that fly segments alternate in their molecular substrates, like the floral wallpaper to the right.* It seems excessively complicated. Furthermore, when we look at the molecular circuitry of segmentation, it's surprising inelegant — each stripe is hard-coded into the regulatory controls of the genes involved. The ugliness of evolutionary contingency is on full display here.
When we look at vertebrates, on the other hand, we see something very different. Segments form sequentially, one at a time from front to back, rather than nearly simultaneously. What we see here is a clock-and-wavefront model in operation, where cyclic surges in the expression of two molecules, fgf8 and Wnt3a, trigger a small pool of cells at the front of an undifferentiated band to pinch off and differentiate into a segment. Then the wave recedes, the front of the undifferentiated band matures a little more, and then when the wave rises again, another set of cells are recruited to make the next segment. And so on and on, until the whole band is delineated into an array of segments.
We find many of the same genes seen in flies involved in this process; for instance, a homolog of the Drosophila pair-rule gene hairy is found in vertebrate segmentation. However, vertebrates don't have the alternating pattern of segmentation — it's the same genes used in each.
Now you're thinking that maybe segmentation evolved independently in vertebrates and arthropods, because I've just highlighted all these major differences. But wait! There's a complication!
Most of what we know about arthropod segmentation comes from Drosophila, and I'll say it again: flies are weird, highly derived, extensively modified to a point where it's surprising they have anything in common with their pre-Cambrian ancestors anymore. All that stuff about near-simultaneous formation of the segments seems to be a product of selection for fast development in flies — those are all recent additions to their developmental programs. We may be getting misled by modern additions to improve the efficiency of the fly-making factory, and so we aren't seeing the original arthropod segmentation plan at all.
To do that, we need to look at an insect that doesn't use this highly derived Drosophila mechanism, and that's easy: there are a lot of insects other than flies out there. Tribolium castaneum, the flour beetle, is a popular alternative. These animals do not form their segments all at once, but instead have a growth zone from which segments differentiate sequentially, from front to back…hey! Like a vertebrate! At least, like a vertebrate superficially — do they also use a clock-and-wavefront mechanism, with periodic surges of signaling gene expression, as vertebrates do?
The tentative answer to that question is yes. Recent observations show that some genes do show clock-like changes in the level of expression in the growth zone.
In the Tribolium embryos below, all were stained for RNA of the pair-rule gene Tc-odd, and you can clearly see the nice segmental stripes of blue Tc-odd RNA in the segmented bits. The interesting part is the posterior growth zone (gz), marked with the gray box in the plots of stain intensity. What you should see in these specimens fixed at different times is a changing level of Tc-odd expression in the gz — do enough of these, and you get a good impression of rising and falling levels of Tc-odd expression over time.


Expression of Tc-odd during early germband elongation. (A to F) In situ hybridization for Tc-odd in elongating germbands at stages 3.II to 5.I, in ventral view (for staging, see supporting online material). The growth zone is located posterior to the black arrowheads. White arrowheads mark the posterior edge of the amniotic fold. (Aâ² to Fâ²) Intensity profiles of Tc-odd expression along the anterior-posterior axis of germbands. The intensity of staining was measured on the lateral part of the germbands shown in (A) to (F), encompassing the ectoderm and the amnion (for explanations, see fig. S1). Note the changing profile of Tc-odd expression within the posterior growth zone (gray area) during the emergence of new primary Tc-odd stripes from that zone. During phase I of stripe formation [(C) and (F)], the growth zone has low levels of Tc-odd, except for two lateral spots of expression that mark the initiation of a new cycle of Tc-odd expression. At phase II [(A) and (D)], Tc-odd expression extends through the growth zone, such that a posterior domain of high Tc-odd expression is established. At phase III [(B) and (E)], Tc-odd expression in the posterior-most part of the growth zone is reduced to low levels, whereas cells at the anterior of the growth zone still express high levels, establishing a broad primary stripe. In a typical collection of embryos, each phase is found in roughly equal numbers. P1 to P4, primary stripes; S1 to S4, intercalating secondary stripes; gz, growth zone.
That isn't a perfect experiment on its own, though. There's variability in the degree of expression of Tc-odd from animal to animal and experiment to experiment, and timing the embryos is tricky and with the possibility of error; what you really want to do is see the changing expression in living embryos. They don't have that assay in Tribolium, yet, but the investigators did something clever instead. They cut embryos in half.
The bifurcated embryos continue developing normally for some hours afterwards, so what you can do is cut them in half, fix one half immediately, let the second half continue developing for some time afterwards, fix it, and then stain both for Tc-odd. Now you can make a direct comparison of the change in gene expression in the same embryo at two different times in development.

(D and Dâ²) In situ hybridization for Tc-odd on two halves of the same embryo. One half was fixed immediately after dissection (D), whereas the other was cultured for 30 min at 30°C before fixation (Dâ²). Tc-odd expression in each half-embryo is at phase 3.II and 3.III, respectively. (E and Eâ²) Same experiment, with half-embryos at phases 4.I and 4. II. The lateral part of the growth zone, encompassing the ectoderm and amnion, is outlined with a dashed line; the medial part, which also includes mesoderm, is marked M (see fig. S1). In panel Eâ², the oblique stripe of Tc-odd expression (white arrowhead) is in the amnion. Anterior is toward the top in all panels.
It's clear. There is a kind of segmentation clock running in Tribolium, and so now you might be leaning back towards thinking it's reasonable to assume that this mechanism might have been inherited from the last common ancestor of both vertebrates and arthropods.
But hang on, we still have unresolved issues. Tribolium also exhibits the alternating segment periodicity we see in Drosophila, but don't see in vertebrates. In fact, when you look at the whole network of segmentation genes in Tribolium it does still look a lot like the fly segmentation network…but not so much like the vertebrate network.
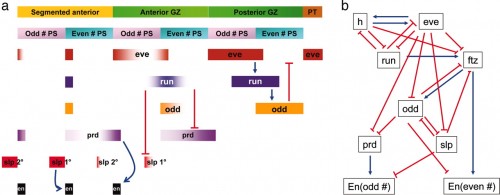
Pair-rule patterning in Tribolium. (a) The dynamic expression of the primary and secondary pair-rule genes and their regulatory interactions are summarized. The bar at the top indicates that anterior is to the left. Newer segments forming in the growth zone are to the right. In this model of pair-rule patterning in Tribolium, two-segment units are prepatterned in the posterior region of the growth zone through one cycle of the regulatory circuit (Tc-eve, Tc-run, and Tc-odd). As the expression of Tc-run retracts anteriorly in even-numbered parasegments, the expression of Tc-prd is derepressed. Primary Tc-prd stripes resolve into two secondary stripes, showing alternatively weak and strong segmental expression. The strong secondary stripes in odd-numbered parasegments regulate Tc-En expression. Tc-run also retracts posteriorly in odd-numbered parasegments, resulting in derepression of the primary Tc-slp stripes. As Tc-run expression fades, expression of the primary Tc-slp stripe extends to the posterior border of the odd-numbered parasegment, which is required for the activation of Tc-En. GZ, growth zone; PS, parasegment; PT, posterior tip. (b) The more complex pair-rule network in Drosophila.
Aaargh, so maybe segmentation evolved independently in insects and vertebrates after all?
But wait again…there are big differences between Tribolium and Drosophila. Genes interact differently, and different genes have subtly different roles in setting up the pattern.
In Drosophila, the three primary pair-rule genes (h, eve, and run) are key players in initiating pair-rule patterning. However, Tc-h seems not to function as a pair-rule gene at all. Although odd is a secondary pair-rule gene in Drosophila that is repressed by eve, Tc-odd functions as a primary pair-rule gene in Tribolium that represses Tc-eve. Repression of slp and odd by eve is critical to activate prd-dependent odd-numbered and ftz-dependent even-numbered en stripes, respectively, in Drosophila In contrast, Tc-eve is required for the activation of Tc-odd, which in turn represses Tc-eve to prepattern a two-segment unit. Furthermore, Tc-run, which is induced by Tc-eve, is important for the formation of Tc-prd-dependent odd-numbered and Tc-slp-dependent even-numbered Tc-en stripes. Drosophila ftz is a secondary pair-rule gene that activates even-numbered en stripes, but Tc-ftz does not function in segmentation. Differences in the primary pair-rule genes result in different genetic interactions between primary and secondary genesand likely affect the regulatory interactions between pair-rule and segment polarity genes. For example, loss of slp affects odd- numbered parasegments, whereas loss of Tc-slp affects even-numbered parasegments.
What this tells me is that this tangled web of interacting gene products is robust and flexible; it can be tweaked by evolutionary changes and still function to produce a reliable segmental pattern. It's also the kind of module that has a core circuit, and has accumulated lots of supplemental inputs that can reinforce the segmental pattern generation, and can also be swapped in for core elements — flies use h-eve-run as the primary patterning genes, while beetles use eve-run-odd, so the actors can shift about in their roles while still producing a recognizably similar play.
Also, these similar networks can function in very different settings: I'm surprised to see the same core players in the all-at-once segmentation pattern in flies and the one-at-a-time sequential segmentation of beetles. What we need is much more comparative molecular data on segmentation networks so we can identify the minimal segmentation core and how it works. Then we have a chance of determining when that basic module evolved, and where it belongs in phylogeny.
Until then, we don't know whether segmentation is a synapomorphy or a homoplasy between vertebrates and arthropods, which is actually kind of cool. Mysteries in science are waiting to be resolved, and the data is on the way!
Choe CP, Miller SC, Brown SJ (2006) A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc Nat Acad Sci USA 103(17):6560-6564.
Sarrazin AF, Peel AD, Averof M (2012) A segmentation clock with two-segment periodicity in insects. Science 336:338-341.
*Yeah, that's the hideous wallpaper in my bedroom, put there in the 1950s, I think. My only excuse is that almost all the time I spend in that room either it's dark or my eyes are closed.
- Log in to post comments



