Nick's post on Amantadine resistance in swine flu was so interesting, I had to look at the protein structures myself.
I couldn't find any structures with the S31N mutation that Nick discussed, but I did find some structures with the M2 protein and Amantadine. Not only are these structures beautiful, but you can look at them and see how the protein works and how the drug prevents the protein from functioning.
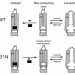
As Nick mentions, the M2 protein from influenza makes a channel for hydrogen ions within the viral membrane. The channel controls the pH inside the virus by opening and closing.
When the channel is open (top row), hydrogen ions pass through the channel and drop the pH. This makes the interior more acidic and activates some of the viral proteins, a necessary step in viral reproduction.
When the channel is closed (middle row), the passage of ions is blocked and the pH remains stable.
One other interesting feature is the location of charged and uncharged residues. I used Cn3D to color the structures by charge. This makes the positively charged amino acids (lysine, arginine, histidine) appear blue and the negatively charged amino acids (aspartic acid and glutamic acid), appear red. Everything else is gray. The pictures below show faces of the protein that are on the outside or inside of a membrane. Those faces show charged amino acids. Around the outside of the protein though, are uncharged amino acids. This is cool because that's what we would expect, hydrophobic or uncharged amino acids are those facing the interior of the lipid membrane.



- Log in to post comments


Coolio!
Thanks for the shout-out, and nice post. The mechanism of inhibition of M2 inhibition by adamantane drugs is still very controversial, though. One structure (the one you used in the post) found the drug bound in the channel, but another found it bound outside of the channel, hinting at an allosteric mechanism of inhibition. Both groups provide some evidence for the validity of their structures, so it'll be interesting to see how this scientific controversy resolves itself in the future.
This just made my day that much brighter. Thanks a million. Something else I stumbled
across was this http://www.drugdelivery.cam. Take a look!
Keep up the great work!
This just made my day that much brighter. Thanks a million. Something else I stumbled
across was this http://www.drugdelivery.cam. Take a look!
Keep up the great work!